LINC00473/miR-374a-5p regulates esophageal squamous cell carcinoma via targeting SPIN1 to weaken ...
LINC00473/miR-374a-5p regulates esophageal squamous cell carcinoma via targeting SPIN1 to weaken the effect of radiotherapy
WILEY journal Cellular Biochemistry
Weizuo Chen1, Yanshan Zhang1, Huijuan Wang2, Tingting Pan1, Yinguo Zhang3, Chao Li3
1 Department of Radiotherapy, Tumor Hospital of Wuwei, Wuwei, Gansu, China
2 Department of Tumor Chemotherapy, Tumor Hospital of Wuwei, Wuwei, Gansu, China
3 Department of Thoracic Surgery, Tumor Hospital of Wuwei, Wuwei, Gansu, China
Abstract
Esophageal squamous cell carcinoma (ESCC) is the most prevalent type in esophageal cancers. Despite accumulating achievements in treatments of ESCC, patients still suffer from recurrence because of treatment failures, one of the reasons for which is radioresistance. Therefore, it is a necessity to explore the molecular mechanism underlying ESCC radioresistance. Long intergenic noncoding RNA 473 (LINC00473) has been reported to be aberrantly expressed in several human malignancies. However, its biological function in radiosensitivity of ESCC remains to be fully understood. This study explored the role of LINC00473 in radiosensitivity of ESCC cells and whether LINC00473 acted as a competing endogenous RNA to realize its modulation on radioresistance. We found that LINC00473 was markedly upregulated in ESCC tissues and cell lines, and its expression was remarkably related to cellular response to irradiation. In addition, knockdown of LINC00473 could sensitize ESCC cells to radiation in vitro. As for the underlying mechanism, we uncovered that there was a mutual inhibition between LINC00473 and miR-374a-5p. Spindlin1 (SPIN1) was verified as a downstream target of miR-374a-5p, and LINC00473 upregulated SPIN1 expression through negatively modulating miR-374a-5p expression. Furthermore, we revealed that SPIN1 could aggravate the radioresistance of ESCC cells. Finally, overexpression of SPIN1 reversed the LINC00473 silencing-enhanced radiosensitivity in ESCC cells. To sum up, we demonstrated that LINC00473 facilitated radioresistance by regulating the miR-374a-5p/SPIN1 axis in ESCC.
KEYWORDS
esophageal squamous cell carcinoma, LINC00473, miR-374a-5p, radiosensitivity, SPIN1
1. INTRODUCTION
Esophageal cancer is the eighth most aggressive malignancy and the sixth commonest reason for cancer-related mortality around the world, featuring with poor prognosis with less than 20% 5-year overall survival (OS) in view of early metastasis.1-3 In China, esophageal squamous cell carcinoma (ESCC) takes about 88.8% of all esophageal cancers.4 Although radiotherapy is widely applied therapy for patients suffering from an unresectable esophageal tumor, the increased radioresistance in recurrent tumors becomes one of the major causes of treatment failure.5,6
Long noncoding RNAs (lncRNAs), a type of mature RNAs with the transcription length of above 200 bases, are discovered by mounting studies to be able to act as competing endogenous RNAs (ceRNAs) contending with downstream target genes for microRNAs (miRNAs).7,8 They are reported to be closely related to tumor proliferation, metastasis, invasion, as well as prognosis, bearing great potential as novel targets for cancer therapy.9,10 Also, the participation of lncRNAs in ESCC has been documented.9,11,12 The oncogenic role of LINC00473 has been identified in numerous cancers.13-16 However, its regulatory function in ESCC remains elusive, so does its role in cell resistance to radiotherapy for cancers, including ESCC.
miRNAs, recognized as a class of endogenous small noncoding RNAs at 19 to 25 nucleotides of length, can regulate the expression of their target genes via targeting the 3ʹ-untranslated region (3′UTR) of messenger RNAs, resulting in translational repression or degradation of mRNAs.17 Among them, downregulation of miR-374a-5p has been discovered in various cancers.18-20 Nevertheless, its role in ESCC remains to be investigated.
Spindlin1 (SPIN1), belonging to SPIN/SSTY family, was initially discovered to be highly expressed in ovarian cancer.21 The oncogenic role of SPIN1 has been subsequently validated by increasing research. For instance, upregulation of SPIN1 induces transformation and proliferation of NIH3T3 cells.22 To date, its oncogenic function has been identified in multiple cancers.23-27 However, neither has its regulatory function nor its relation with radioresistance been explored in ESCC yet.
The present study aimed to explore the role of LINC00473/miR-374a-5p/SPIN1 in radiotherapy for ESCC.
2. MATERIALS AND METHODS
2.1 Tissue samples
Ninety-six matched human ESCC tissues and corresponding nontumor tissues were collected after direct surgical resection performed at the Tumor Hospital of Wuwei. All of the tissue samples from patients were immediately frozen in liquid nitrogen and stored at -80°C for later use. Our research protocol was approved by the Tumor Hospital of Wuwei. Standard written consent was obtained from each patient.
2.2 Cell lines and cell culture
Human ESCC cell lines (TE-1, EC9706, Eca109, and Kyse450) and human normal human esophageal epithelial cell line (HET-1A) were purchased from the Shanghai Institute of Biochemistry and Cell Biology (Shanghai, China). All of the cell lines were maintained in Roswell Park Memorial Institute (RPMI)-1640 medium (Invitrogen, Carlsbad, CA) with 10% fetal bovine serum (Gibco, Gaithersburg, MD) supplemented with 100 U/mL penicillin and 100 μg/mL streptomycin (Invitrogen) in a humidified environment with 5% CO2 at 37°C.
2.3 Quantitative real-time polymerase chain reaction
For quantitative real-time polymerase chain reaction (qRT-PCR), total RNA was reverse-transcribed with reverse transcription kit (Takara, Otsu, Japan) and Bulge-Loop miRNA-specific reverse transcription primers (RiboBio, Guangzhou, China). Quantitative PCR was performed with SYBR Premix Ex Taq (Takara, Dalian, China) reagents and Bulge-Loop primers in an ABI PRISM 7900 System (Applied Biosystems, Carlsbad, CA) with glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and U6 as a normalization control. The primers used were as follows:
5′-GATGGAAAGGAGGGAAGG-3′ (forward) and 5′-CACAGTGGGTCCAGGGTT-3′ (reverse) for LINC00473; 5′-GCGCGCTTATAATACAACCTGA-3′ (forward) and 5′-GTGCAGGGTCCGAGGT-3′ (reverse) for miR-374a-5p; 5′-GGTATCCAGGTGCCTTATG-3′ (forward) and 5′-GGACGATCTGCGACTATTG-3′ (reverse) for SPIN1; 5′-GAAGGTGAAGGTCGGAGT-3′ (forward) and 5′-GATGGCAACAATATCCACTT-3′ (reverse) for GAPDH; and 5′-CTCGCTTCG GCAGCACA-3′ (forward) and 5′-AACGCTTCACGAATTT GCGT-3′ (reverse) for U6.
2.4 Cell transfection
For cell transfection assay, pcDNA3.1 subcoloned with LINC00473 or short hairpin RNAs (shRNAs) specifically targeting LINC00473 were used to overexpress or knockdown LINC00473, and pcDNA3.1 inserted with the full sequence of SPIN1 or shRNAs targeting against SPIN1 were used to overexpress or knockdown SPIN1, with pcDNA3.1 or sh-NC as a negative control (NC). And miR-374a-5p-mimic or miR-374a-5p-inhibitor were used to overexpress or knockdown miR-374a-5p with NC-mimic or NC-inhibitor as a negative control. All vectors were purchased from Shanghai Integrated Biotech Solutions Co, Ltd (Shanghai, China) and transfected as demanded into cells utilizing Lipofectamine 2000 (Invitrogen) under manufacturer's protocols. The efficiency of transfection was evaluated by qRT-PCR.
2.5 Cell Counting Kit-8
TE-1 and EC9706 cells, with a density of 2 × 10³ cells per well, were seeded in 96-well plates. Cell viability was measured using Cell Counting Kit-8 (CCK-8; Beyotime, Haimen, China). Following incubation for 0, 24, 48, 72, and 96 hours, 10 μL of CCK-8 solution was supplemented to each well of the 96-well plate and incubated for 2 hours in an incubator. Following incubation, phosphate-buffered saline was utilized to wash the plates. The absorbance was read at a wavelength of 450 nm, and a proliferation curve according to the absorbance and time was constructed.
2.6 Colony survival assay
Transfected TE-1 and EC9706 cells were seeded into six-well plates and exposed to the indicated single doses of irradiation (0, 2, 4, 6, or 8 Gy). Subsequent to 2 weeks of incubation at 37°C, cells were fixed with 100% methanol and stained by 0.1% crystal violet in absolute ethanol for 15 minutes. The visible colonies with more than 50 cells were calculated with an inverted microscope. The survival fraction (SF) was counted as the ratio of plating efficiency for irradiated and nonirradiated cells. Plating efficiency was defined as below: the number of colonies divided by the number of cells seeded × 100%. The data were fitted using a classic single-hit multitarget model: y = 1-(1-e-D/D0)N to draw cell survival curves. Radiation-associated parameters, including D0 (mean lethal dose), Dq (quasi-threshold dose), and N (extrapolation number), were calculated according to the curve. The sensitivity enhancement ratio is presented as the ratio of control group D0 value to treatment group D0 value.
2.7 Luciferase reporter
The wt-LINC00473 or wt-SPIN1 with the binding sequences on the 3′-UTR of LINC00473 or SPIN1, and the mut-LINC00473 or mut-SPIN1 with the mutant sites were synthesized in vitro based on bioinformatics predictions. Then NC-mimic or miR-374a-5p-mimic was transfected with wt-LINC00473 or mut-LINC00473 luciferase reporter plasmids, and NC-mimic or miR-374a-5p-mimic or miR-374a-5p-mimic plus pcDNA3.1-LINC00473 were transfected with wt-SPIN1 or mut-SPIN1 luciferase reporter plasmids, respectively, into TE-1 and EC9706 cells by Lipofectamine 2000 Reagents (Invitrogen) according to the manufacturer's protocol. Thirty-eight hours post-transfection, luciferase activity was detected using the Dual-Luciferase Reporter Assay System (Promega Corporation, Madison, WI). Renilla luciferase activities were used for normalization.
2.8 Western blot analysis
Total cell extracts were obtained and the proteins were isolated using RIPA lysis buffer (Beyotime Biotechnology, China) together with protease inhibitors (Roche, Shanghai, China). Then the protein was quantified with BCA Protein Assay Kit (Pierce, Appleton, WI) and electrophoresed by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis. After blotting the protein onto polyvinylidene difluoride membranes (Millipore, Bedford, MA), the membranes were sealed by skim milk. The membranes were then cultivated at 4°C under room temperature with primary antibodies overnight and later with secondary antibody for another 2 hours. Primary antibodies were: anti-PARP (Santa Cruz, CA), anti-SPIN1 (Sangon Biotech, Shanghai, China), and anti-GAPDH (Cell Signaling Technology, Danvers, MA) as a loading control. Signals were detected using ECL Kit (Amersham Pharmacia Biotech, Piscataway, NJ). Images were scanned by FujiFilm LAS-1000 (FujiFilm, Düsseldorf, Germany).
2.9 Statistical analysis
Data were presented as mean ± SD from at least three independent experiments. Statistical analyses were performed with SPSS 18.0 Software (SPSS Inc, Chicago, IL). The difference between two groups was analyzed using a two-tailed Student's t-test and analysis of variance followed by Student-Newman-Keuls Q test was used for multigroup comparison. The expression correlation was explored by Spearman's correlation analysis. Kaplan-Meier analysis as well as log-rank test were carried out to examine the association of LINC00473 and SPIN1 with ESCC. Significant associations between LINC00473 level and clinico-pathological parameters were assessed using a χ² test. Two-sided P < 0.05 was considered to be statistically significant.
3. RESULTS
3.1 Upregulation of LINC00473 indicated radioresistance and poor prognosis in ESCC patients
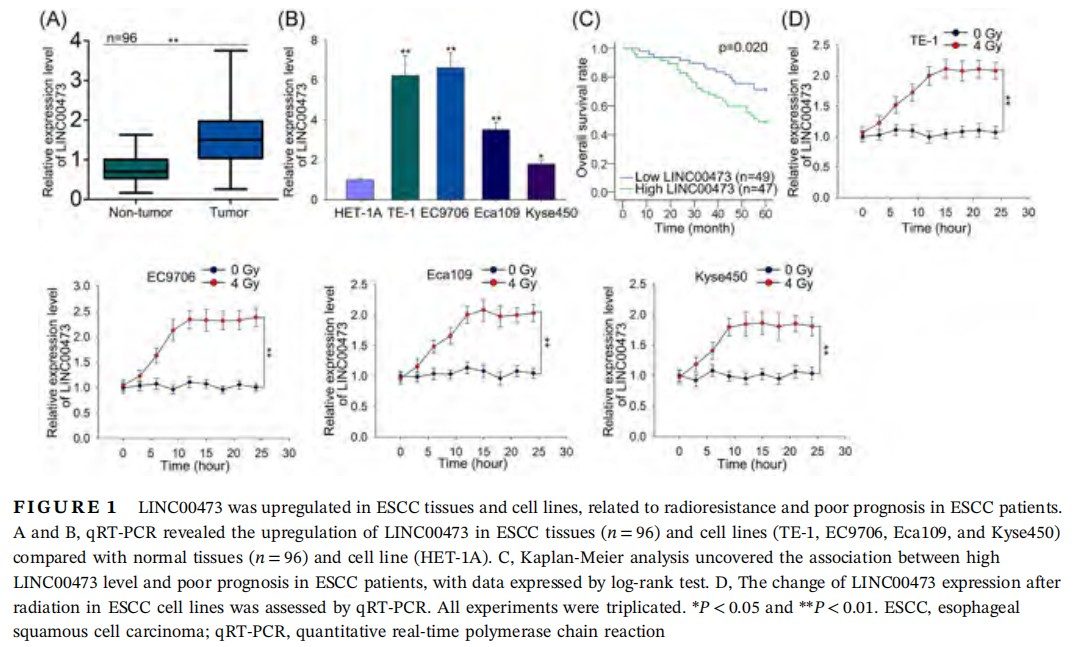
To begin with, to investigate whether LINC00473 was involved in ESCC, we examined its expression in tissues and cell lines. qRT-PCR results demonstrated a significantly high level of LINC00473 in ESCC tumor tissues (n = 96) and ESCC cell lines (TE-1, EC9706, Eca109, and Kyse450) compared with corresponding normal tissues (n = 96) and normal human esophageal mucosal epithelial cell line (HET-1A) (Figure 1A and 1B). Notably, TE-1 and EC9706 cells presented the highest LINC00473 level among all ESCC cell lines (Figure 1B). Also, by examining the relation of LINC00473 with clinical characteristics of ESCC, we found that LINC00473 was closely related to N stage (P = 0.011), M stage (P = 0.001), and TNM stage (P = 0.021) (Table 1). And the independent prognostic significance of LINC00473 (P = 0.036) in ESCC patients was confirmed as shown in Table 2. What's more, Kaplan-Meier analysis revealed that high LINC00473 level in ESCC patients led to poor prognosis (Figure 1C). To probe the association of LINC00473 with ESCC cell radioresistance, we examined the expression change of LINC00473 in four ESCC cell lines after 4 Gy radiation, finding that LINC00473 was upregulated after 4 Gy radiation (Figure 1D). The above results suggested the pro-radioresistance potential of LINC00473, as well as its prognostic significance in ESCC patients.
3.2 Knockdown of LINC00473 inhibited proliferation and promoted radiosensitivity of ESCC cells
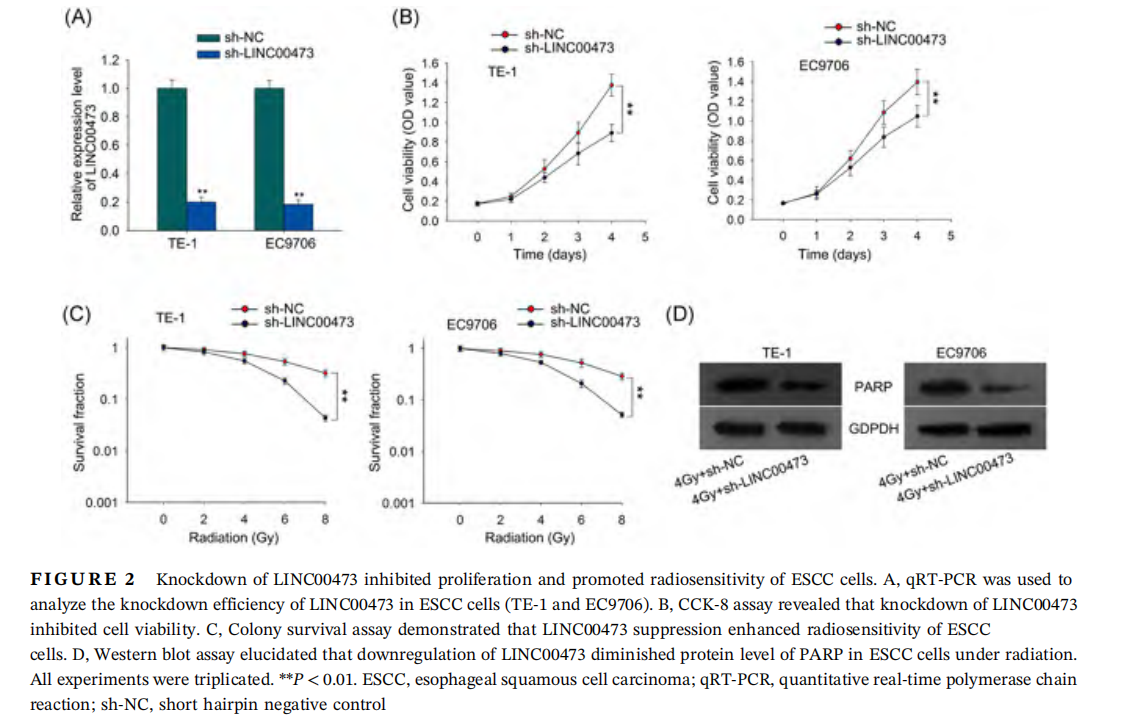
We investigated the biological role of LINC00473. We silenced LINC00473 using specific shRNAs targeting LINC00473 (shLINC00473), which caused obvious knockdown of LINC00473 in TE-1 and EC9706 cells (Figure 2A). Since we discovered that LINC00473 expression was related to TNM stage in ESCC patients as presented in Table 1, indicating that LINC00473 might be associated with tumor progression in ESCC, and cell proliferation is an important cellular process in tumor progression, we first detected the effect of LINC00473 on cell proliferation by CCK-8 assay. It was illustrated in Figure 2B that downregulation of LINC00473 attenuated proliferation of TE-1 and EC9706 cells compared with the control group in both cell lines. Next, we interrogated the effect of LINC00473 on cell radioresistance. Colony survival assay depicted that with the accumulated dose of radiation, suppression of LINC00473 level caused a lower SF in both cell lines (Figure 2C), indicating the enhancement of ESCC cell sensitivity to irradiation under LINC00473 silencing. Additionally, we detected the level of poly-(ADP-ribose) polymerase (PARP), a protein responsible for DNA damage repair, after treating both cell lines with 4 Gy dose of radiation. Western blot analysis revealed that knockdown of LINC00473 led to a reduction of PARP protein level (Figure 2D), suggesting that suppressing LINC00473 inhibited DNA repair under radiation to promote radiosensitivity of ESCC cells. These results suggested that knockdown of LINC00473 inhibited proliferation and promoted radiosensitivity of ESCC cells.
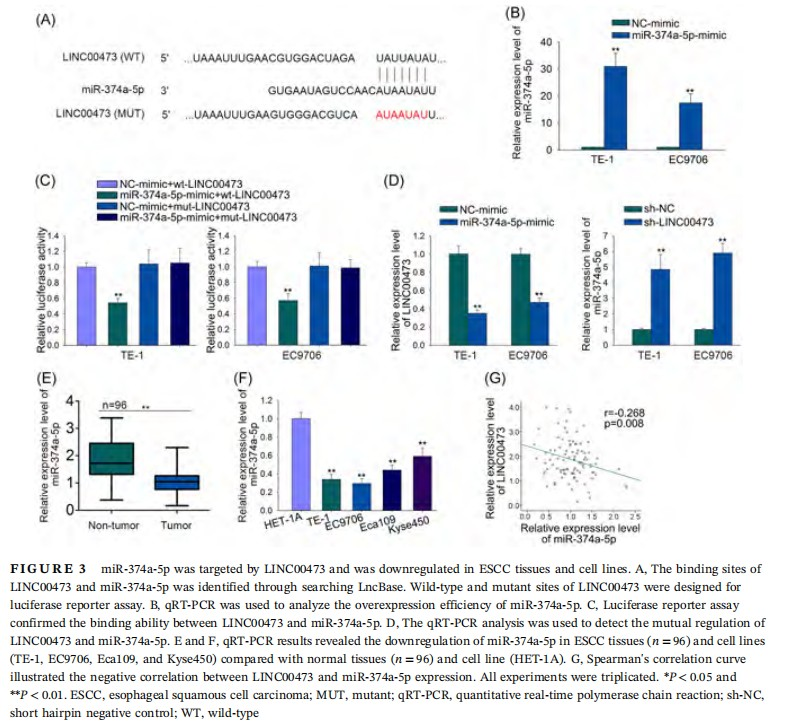
3.3 miR-374a-5p was targeted by LINC00473 and downregulated in ESCC tissues and cell lines
After confirming the inductive effect of LINC00473 on radioresistance in ESCC cells, we further investigated its underlying regulatory mechanism. It has been known that lncRNAs can act as ceRNAs to bind with target miRNAs.7,8 Therefore, we speculated that LINC00473 might regulate radiosensitivity of ESCC cells via this mechanism. By searching LncBase (http://carolina.imis.athena-innovation.gr/diana_tools/web/index.php?r=lncbasev2%2Findex-predicted), we identified a number of potential binding miRNAs for LINC00473. Among them, we selected miR-374a-5p because it has been discovered to be downregulated in colorectal cancer and melanoma, but its regulatory mechanism in cancers is rarely explored, including in ESCC.20,28-30 The binding sites between LINC00473 and miR-374a-5p and the mutant sites were shown in Figure 3A. The transfection of miR-374a-5p-mimic into ESCC cells was confirmed by qRT-PCR analysis (Figure 3B). To investigate the binding ability between LINC00473 and miR-374a-5p, wild-type or mutant LINC00473 reporter plasmids were constructed and cotransfected with miR-374a-5p-mimic or NC-mimic into two ESCC cell lines (TE-1 and EC9706). Luciferase reporter assay revealed that overexpression of miR-374a-5p weakened the luciferase activity of wt-LINC00473 rather than mut-LINC00473 (Figure 3C), confirming the binding ability between LINC00473 and miR-374a-5p. Additionally, the qRT-PCR analysis revealed that miR-374a-5p upregulation decreased LINC00473 level, and vice versa (Figure 3D). Then, we examined the association of miR-374a-5p with ESCC by detecting its level in tissues and cell lines. It was shown by qRT-PCR results that miR-374a-5p was significantly downregulated in ESCC tissues (n = 96) and cell lines (TE-1, EC9706, Eca109, and Kyse450) (Figure 3E and 3F). Spearman's correlation curve revealed the negative correlation between LINC00473 and miR-374a-5p in ESCC tissues (Figure 3G). These results indicated that miR-374a-5p was targeted by LINC00473 and was downregulated in ESCC tissues and cell lines.
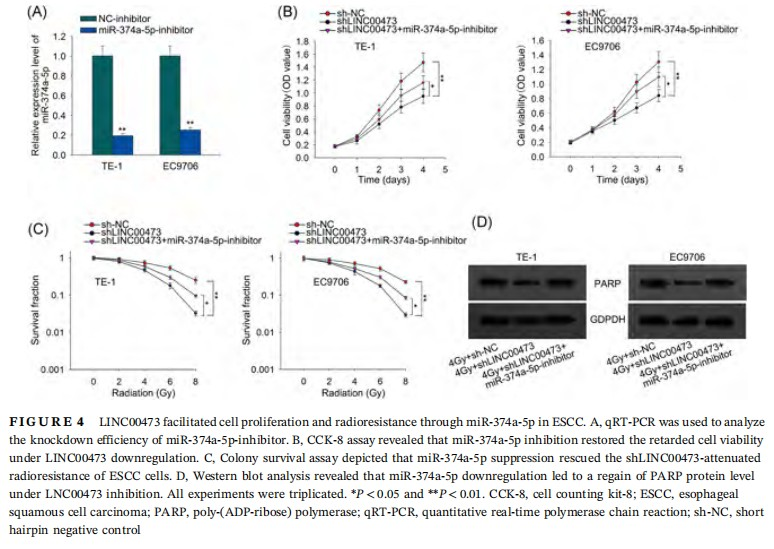
3.4 LINC00473 facilitated cell proliferation and radioresistance through miR-374a-5p in ESCC
To test whether LINC00473 regulated radiosensitivity of ESCC cell through miR-374a-5p, we downregulated miR-374a-5p using miR-374a-5p inhibitors for rescue assays. The pronounced downregulation of miR-374a-5p under the transfection of miR-374a-5p-inhibitor was confirmed by qRT-PCR (Figure 4A). CCK-8 assay demonstrated that knockdown of miR-374a-5p rescued the retarded cell viability under LINC00473 downregulation (Figure 4B). Colony survival assay revealed that miR-374a-5p inhibition restored the reduction of colony SF of ESCC cells caused by LINC00473 knockdown under accumulated radiation dose (Figure 4C). Also, Western blot analysis results illustrated that knockdown of miR-374a-5p caused a regain of PARP protein level which was diminished by LINC00473 silence under the treatment of radiation (Figure 4D). Together, the results implied that LINC00473 facilitated cell proliferation and radioresistance through miR-374a-5p in ESCC.
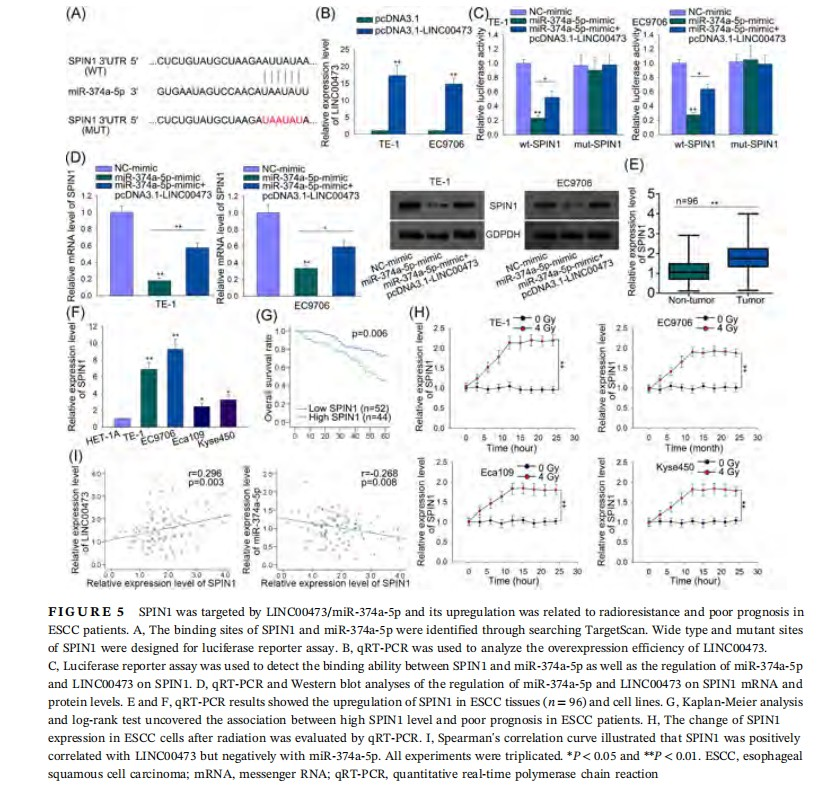
3.5 SPIN1 was targeted by LINC00473/miR-374a-5p, and its upregulation predicted radioresistance and poor prognosis in ESCC
It has been known that miRNAs can regulate gene expression by directly binding with the 3ʹUTR of their target genes.17 Thus, we searched TargetScan (http://www.targetscan.org/vert_72/) for the potential downstream target genes for miR-374a-5p. Among all potential interacting partners for miR-374a-5p, SPIN1 has been reported to be an oncogene in multiple cancers.23-27 However, its role in ESCC as well as its effect on radiotherapy remain unclear. Hence, we chose SPIN1 for further investigation.
To examine whether SPIN1 competed with LINC00473 for miR-374a-5p, we constructed luciferase reporter plasmids with wild-type or mutant SPIN1 3ʹUTR. The wild-type and mutant binding sequences of SPIN1 with miR-374a-5p were shown in Figure 5A. And pcDNA3.1-LINC00473 was transfected into ESCC cells, causing a significant overexpression of LINC00473 as analyzed by qRT-PCR, with pcDNA3.1 vector as negative control (Figure 5B). Luciferase reporter assay showed that in both cell lines, miR-374a-5p overexpression led to a reduction of luciferase activity on wild-type SPIN1, and cotransfection of pcDNA3.1-LINC00473 recovered the reduction, but no obvious variation of luciferase activity was detected on mut-SPIN1 (Figure 5C). Additionally, qRT-PCR and Western blot analysis elicited that upregulation of miR-374a-5p decreased SPIN1 mRNA and protein levels, while the cotransfection with pcDNA3.1-LINC00473 reversed this decrease (Figure 5D). These results validated that SPIN1 competed with LINC00473 to bind with miR-374a-5p.
After that, we investigated the association between SPIN1 and ESCC. qRT-PCR results showed that SPIN1 was upregulated in ESCC tumor tissues (n = 96) and cell lines compared with normal ones (Figure 5E and 5F). Kaplan-Meier analysis revealed the positive relation of high SPIN1 level with poor outcome in ESCC patients (Figure 5G). Also, the detection on the change in SPIN1 level in ESCC cell lines after radiation revealed that SPIN1 might associate with ESCC cell radioresistance (Figure 5H). Finally, Spearman's correlation curve elucidated that SPIN1 was positively correlated with LINC00473 whereas negatively correlated with miR-374a-5p expression (Figure 5I). Together, these results verified that SPIN1 competed with LINC00473 for miR-374a-5p and its upregulation was related to radioresistance and poor prognosis in ESCC patients.
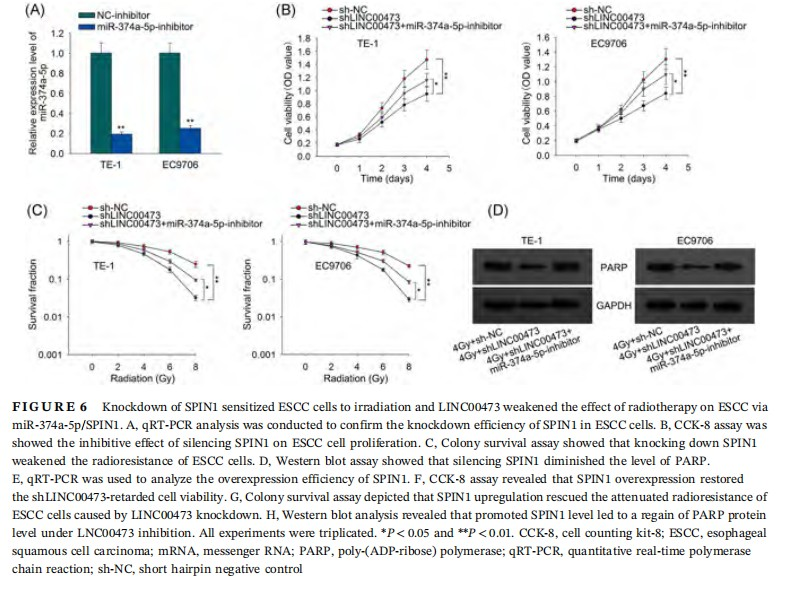
3.6 Knockdown of SPIN1 sensitized ESCC cells to irradiation and LINC00473 weakened the effect of radiotherapy on ESCC via miR-374a-5p/SPIN1
Since the role of SPIN1 in ESCC cell response to irradiation has never been probed, we first unfolded function assays to explore the influence of SPIN1 on ESCC cell radiosensitivity. qRT-PCR results confirmed the knockdown of SPIN1 by sh-SPIN1 transfection in ESCC cells (Figure 6A). We observed through CCK-8 assay that silencing SPIN1 lowered cell proliferation in ESCC cells (Figure 6B). Colony survival assay depicted that the SF of SPIN1-silenced ESCC cells presented a sharper decrease with the accumulated dose of irradiation (Figure 6C). And Western blot analysis revealed that knockdown of SPIN1 diminished the protein levels of PARP in ESCC cells (Figure 6D). Finally, to investigate whether LINC00473/miR-374a-5p regulated radiosensitivity of ESCC cells by targeting SPIN1, we overexpressed SPIN1 for rescue assays. As presented in Figure 6E, SPIN1 was obviously overexpressed after the transfection of pcDNA3.1-SPIN1 in TE-1 and EC9706 cells. CCK-8 assay demonstrated that overexpression of SPIN1 rescued the LINC00473 downregulation-decreased cell viability (Figure 6F). Colony survival assay revealed that SPIN1 upregulation restored the reduction of colony SF of ESCC cells caused by LINC00473 knockdown under accumulated radiation dose (Figure 6G). Also, Western blot analysis results illustrated that with the treatment of radiation, inducing SPIN1 caused a regain of LINC00473 overexpression-lessened PARP protein level (Figure 6H). To sum up, the results above proved that LINC00473 weakened the effect of radiotherapy on ESCC via miR-374a-5p/SPIN1.
4. DISCUSSION
ESCC is the most prevalent type of esophageal cancer, with poor prognosis and high reoccurrence rate among patients.1-4 Despite the improvement of radiotherapy, which is one of the primary options for patients with unresectable tumors, the reoccurrence rate remains dismal because of the radioresistance.5,6 It has been reported that lncRNAs participate in multiple cancers and regulate radioresistance of cancer cells, including ESCC.9-12 LINC00473 has been discovered to be a tumor promoter in several cancers.13-16 Accordingly, the present study firstly revealed the role of LINC00473 in the resistance to radiotherapy for ESCC. To begin with, we discovered the significant upregulation of LINC00473 in ESCC tumor tissues and cell lines, and confirmed the association of LINC00473 with radioresistance and its prognostic significance in ESCC patients. Loss-of-function assays indicated the pro-radioresistance role of LINC00473 in ESCC.
microRNAs (miRNAs) have been reported to be a group of endogenous small noncoding RNAs that are 19 to 25 bases long, modulating gene expression via targeting the 3′UTR of mRNAs and thus contributing to translational repression or degradation of mRNAs.17 Previous studies have reported their involvement in ceRNA network, competed by lncRNAs and downstream target genes.7,8 Present study confirmed that LINC00473 regulated radioresistance of ESCC cells through ceRNA mechanism. By searching LncBase, we selected miR-374a-5p from various potential binding partners for LNC00473 because it has been reported to exert tumor-suppressive roles in many other cancers, while its role in ESCC remains uncovered.13-16 Present study firstly validated the interaction between LINC00473 and miR-374a-5p as well as their negative correlation in ESCC tissues. Later, rescue assays proved that LINC00473 regulated ESCC cell radiosensitivity through miR-374a-5p.
SPIN1 is known to be a member in SPIN/SSTY family. Its upregulation was first discovered in ovarian cancer.21 Later, the role of SPIN1 as a tumor promoter has been validated by other studies such as the discovery that upregulation of SPIN1 induces transformation and proliferation of NIH3T3 cells.22 To date, its oncogenic function has been identified in multiple cancers.23-27 However, both its regulatory function in ESCC as well as its relation with radioresistance in cancer have not been explored yet. The present study identified SPIN1 as a downstream target gene for miR-473a-5p via searching TargetScan, and confirmed its involvement in the ceRNA network with LINC00473/miR-374a-5p. What's more, the prognostic and therapeutic significance of SPIN1 were uncovered in ESCC patients. Loss-of-function assays indicated that SPIN1 could enhance the radioresistance of ESCC cells. Finally, rescue assays proved that LINC00473 contributed to radioresistance of ESCC cells through miR-374a-5p/SPIN1 pathway.
In conclusion, the present study revealed for the first time that LINC00473 regulated ESCC cell proliferation and radioresistance through miR-374a-5p/SPIN1 axis, which provides a novel prognostic biomarker and a potential therapeutic target for ESCC. However, more assays should be conducted for further consolidation of this mechanism.
ACKNOWLEDGMENT
We express our gratitude to Tumor Hospital of Wuwei for their support.
CONFLICT OF INTERESTS
The authors declare that there are no conflicts of interest.
AUTHOR CONTRIBUTIONS
WC and YZ contributed to article writing, experiment design, and data analysis. HW, TP, YZ, and CL collected materials. All authors proofread the final version of this manuscript.
ORCID
Yanshan Zhang iD http://orcid.org/0000-0002-8705-9843
REFERENCES
1. Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2014;136:E359-E386.
2. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7-30.
3. Domper Arnal MJ, Ferrández Arenas Á, Lanas Arbeloa Á. Esophageal cancer: risk factors, screening and endoscopic treatment in Western and Eastern countries. World J Gastroenterol. 2015;21:7933-7943.
4. Bedford JS. Sublethal damage, potentially lethal damage, and chromosomal aberrations in mammalian cells exposed to ionizing radiations. Int J Radiat Oncol Biol Phys. 1991;21:1457-1469.
5. Chen X, Guo J, Xi R-X, Chang Y-W, Pan F-Y, Zhang X-Z. MiR-210 expression reverses radioresistance of stem-like cells of esophageal squamous cell carcinoma. World J Clin Oncol. 2014;5:1068-1077.
6. Zhou Y, Chu L, Wang Q, et al. CD59 is a potential biomarker of esophageal squamous cell carcinoma radioresistance by affecting DNA repair. Cell Death Dis. 2018;9:887.
7. Yin H, Wang X, Zhang X, et al. Integrated analysis of lncRNA-associated-competing endogenous RNAs as prognostic biomarkers in clear cell renal carcinoma. Cancer Sci. 2018;10:3336-3349.
8. Yin X, Huang S, Zhu R, Fan F, Sun C, Hu Y. Identification of long non-coding RNA competing interactions and biological pathways associated with prognosis in pediatric and adolescent cytogenetically normal acute myeloid leukemia. Cancer Cell Int. 2018;18:122.
9. Liu B, Sun L, Liu Q, et al. A cytoplasmic NF-κB interacting long noncoding RNA blocks IκB phosphorylation and suppresses breast cancer metastasis. Cancer Cell. 2015;27:370-381.
10. Xing Z, Lin A, Li C, et al. LncRNA directs cooperative epigenetic regulation downstream of chemokine signals. Cell. 2014;159:1110-1125.
11. Chen F-J, Sun M, Li S-Q, et al. Upregulation of the long non-coding RNA hotair promotes esophageal squamous cell carcinoma metastasis and poor prognosis. Mol Carcinog. 2012;52:908-915.
12. Zhou XL, Wang WW, Zhu WG, et al. High expression of long non-coding RNA AFAP1-AS1 predicts chemoradioresistance and poor prognosis in patients with esophageal squamous cell carcinoma treated with definitive chemoradiotherapy. Mol Carcinog. 2016;55:2095-2105.
13. Shi C, Yang Y, Yu J, Meng F, Zhang T, Gao Y. The long noncoding RNA LINC00473, a target of microRNA 34a, promotes tumorigenesis by inhibiting ILF2 degradation in cervical cancer. Am J Cancer Res. 2017;7:2157-2168.
14. Wang L, Zhang X, Sheng L, Qiu C, Luo R. LINC00473 promotes the Taxol resistance via miR-15a in colorectal cancer. Biosci Rep. 2018;38:BSR20180790.
15. Zhang W, Song Y. LINC00473 predicts poor prognosis and regulates cell migration and invasion in gastric cancer. Biomed Pharmacother. 2018;107:1-6.
16. Chen Z, Li J-L, Lin S, et al. cAMP/CREB-regulated LINC00473 marks LKB1-inactivated lung cancer and mediates tumor growth. J Clin Invest. 2016;126:2267-2279.
17. Dalmay T. Mechanism of miRNA-mediated repression of mRNA translation. Essays Biochem. 2013;54:29-38.
18. Lian F, Cui Y, Zhou C, Gao K, Wu L. Identification of a plasma four-microRNA panel as potential noninvasive biomarker for osteosarcoma. PLOS One. 2015;10:e0121499.
19. Sim J, Kim Y, Kim H, et al. Identification of recurrence-associated microRNAs in stage I lung adenocarcinoma. Medicine. 2018;97:e10996.
20. Slattery ML, Pellatt AJ, Lee FY, et al. Infrequently expressed miRNAs influence survival after diagnosis with colorectal cancer. Oncotarget. 2017;8:83845-83859.
21. Yue W, Sun LY, Li CH, Zhang LX, Pei XT. Screening and identification of ovarian carcinomas related genes. Ai Zheng. 2004;23:141-5.
22. Gao Y, Yue W, Zhang P, et al. Spindlin1, a novel nuclear protein with a role in the transformation of NIH3T3 cells. Biochem Biophys Res Commun. 2005;335:343-350.
23. Li Y, Ma X, Wang Y, Li G. miR-489 inhibits proliferation, cell cycle progression and induces apoptosis of glioma cells via targeting SPIN1-mediated PI3K/AKT pathway. Biomed Pharmacother. 2017;93:435-443.
24. Wang J-X, Zeng Q, Chen L, et al. SPINDLIN1 promotes cancer cell proliferation through activation of WNT/TCF-4 signaling. Mol Cancer Res. 2012;10:326-335.
25. Chen X, Wang Y-W, Gao P. SPIN1, negatively regulated by miR-148/152, enhances adriamycin resistance via upregulating drug metabolizing enzymes and transporter in breast cancer. J Exp Clin Cancer Res. 2018;37:100.
26. Drago-Ferrante R, Pentimalli F, Carlisi D, et al. Suppressive role exerted by microRNA-29b-1-5p in triple negative breast cancer through SPIN1 regulation. Oncotarget. 2017;8:28939-28958.
27. Fang Z, Cao B, Liao J-M, et al. SPIN1 promotes tumorigenesis by blocking the uL18 (universal large ribosomal subunit protein 18)-MDM2-p53 pathway in human cancer. eLife. 2018;7:e31275.
28. Margue C, Reinsbach S, Philippidou D, et al. Comparison of a healthy miRNome with melanoma patient miRNomes: are microRNAs suitable serum biomarkers for cancer? Oncotarget. 2015;6:12110-12127.
29. Slattery ML, Herrick JS, Mullany LE, et al. An evaluation and replication of miRNAs with disease stage and colorectal cancer-specific mortality. Int J Cancer. 2015;137:428-438.
30. Wang D, Li YJ, Ding N, et al. Molecular networks and mechanisms of epithelial-mesenchymal transition regulated by miRNAs in the malignant melanoma cell line. Hereditas. 2015;37:673-82.
